A potent and selective inhibitor for the modulation of MAGL activity in the neurovasculature.
Kemble, A.M., Hornsperger, B., Ruf, I., Richter, H., Benz, J., Kuhn, B., Heer, D., Wittwer, M., Engelhardt, B., Grether, U., Collin, L.(2022) PLoS One 17: e0268590-e0268590
- PubMed: 36084029
- DOI: https://doi.org/10.1371/journal.pone.0268590
- Primary Citation of Related Structures:
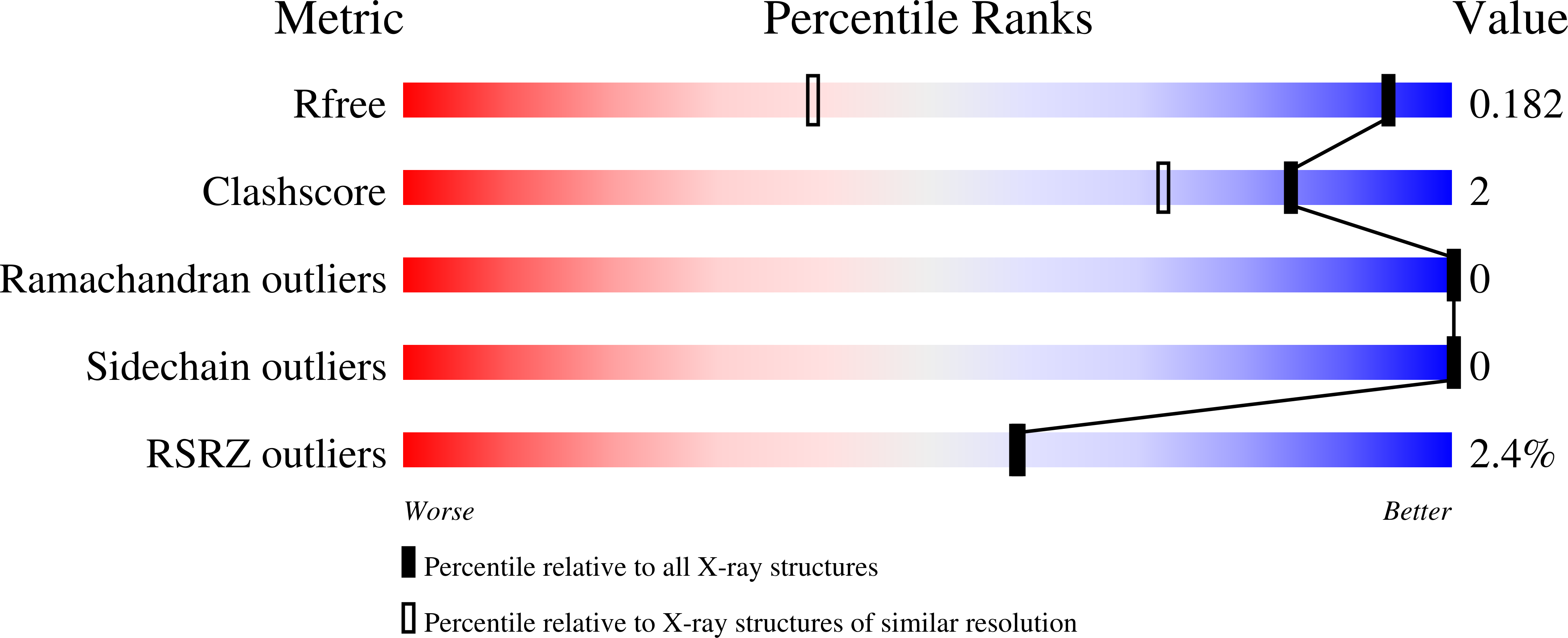
7ZPG - PubMed Abstract:
Chronic inflammation and blood-brain barrier dysfunction are key pathological hallmarks of neurological disorders such as multiple sclerosis, Alzheimer's disease and Parkinson's disease. Major drivers of these pathologies include pro-inflammatory stimuli such as prostaglandins, which are produced in the central nervous system by the oxidation of arachidonic acid in a reaction catalyzed by the cyclooxygenases COX1 and COX2. Monoacylglycerol lipase hydrolyzes the endocannabinoid signaling lipid 2-arachidonyl glycerol, enhancing local pools of arachidonic acid in the brain and leading to cyclooxygenase-mediated prostaglandin production and neuroinflammation. Monoacylglycerol lipase inhibitors were recently shown to act as effective anti-inflammatory modulators, increasing 2-arachidonyl glycerol levels while reducing levels of arachidonic acid and prostaglandins, including PGE2 and PGD2. In this study, we characterized a novel, highly selective, potent and reversible monoacylglycerol lipase inhibitor (MAGLi 432) in a mouse model of lipopolysaccharide-induced blood-brain barrier permeability and in both human and mouse cells of the neurovascular unit: brain microvascular endothelial cells, pericytes and astrocytes. We confirmed the expression of monoacylglycerol lipase in specific neurovascular unit cells in vitro, with pericytes showing the highest expression level and activity. However, MAGLi 432 did not ameliorate lipopolysaccharide-induced blood-brain barrier permeability in vivo or reduce the production of pro-inflammatory cytokines in the brain. Our data confirm monoacylglycerol lipase expression in mouse and human cells of the neurovascular unit and provide the basis for further cell-specific analysis of MAGLi 432 in the context of blood-brain barrier dysfunction caused by inflammatory insults.
Organizational Affiliation:
Roche Pharma Research & Early Development (pRED), Roche Innovation Center Basel, F. Hoffmann-La Roche Ltd., Basel, Switzerland.